Section: New Results
Medical Image Analysis
Longitudinal Analysis and Modeling of Brain Development
Participants : Mehdi Hadj-Hamou [Correspondent] , Xavier Pennec, Nicholas Ayache, Hervé Lemaitre [Inserm U1000] , Jean-Luc Martinot [Inserm U1000] .
This work is partly funded through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Processing pipeline - brain development - adolescence - longitudinal analysis - non-rigid registration algorithm - extrapolation
-
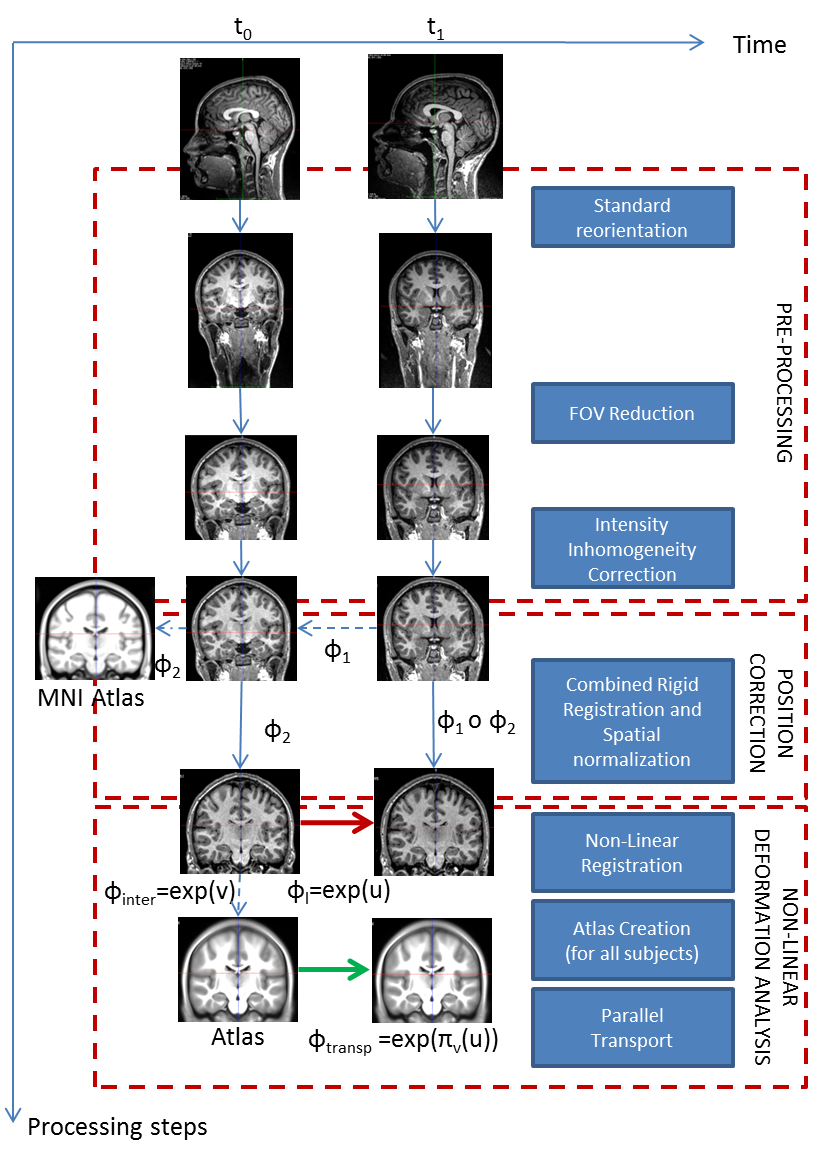
We proposed and detailed a deformation-based morphometry computational framework, called Longitudinal Log-Demons Framework (LLDF), which estimates the longitudinal brain deformations from image data series, transports them in a common space and performs statistical group-wise analyses (see Fig. 1 ). This processing pipeline is based on freely available softwares and relies on the LCC log-Demons non-linear diffeomorphic registration algorithm with an additional modulation of the similarity term using a confidence mask to increase robustness with respect to brain boundary intensity artifacts.
-
The LLDF framework is applied to the study of longitudinal trajectories during adolescence, for which little is known. The aim of this project is to provide models of brain development during adolescence based on diffeomorphic registration parametrised by SVFs. Our study focused particularly on the link between sexual dimorphism and the longitudinal evolution of the brain. This work was done in collaboration with J.L. Martinot et H. Lemaître (Inserm U1000).
|
Inter-Operative Relocalization in Flexible Endoscopy
Participants : Anant Suraj Vemuri [Correspondent] , Stéphane Nicolau, Luc Soler, Nicholas Ayache.
This work has been performed in collaboration with IHU Strasbourg and IRCAD, France.
Computer Assisted Intervention, Barrett's Esophagus, Biopsy Relocalization, Electromagnetic tracking
Oesophageal adenocarcinoma arises from Barrett’s oesophagus, which is the most serious complication of gastro-oesophageal reflux disease. Strategies for screening involve periodic surveillance and tissue biopsies. A major challenge in such regular examinations is to record and track the disease evolution and relocalization of biopsied sites to provide targeted treatments.
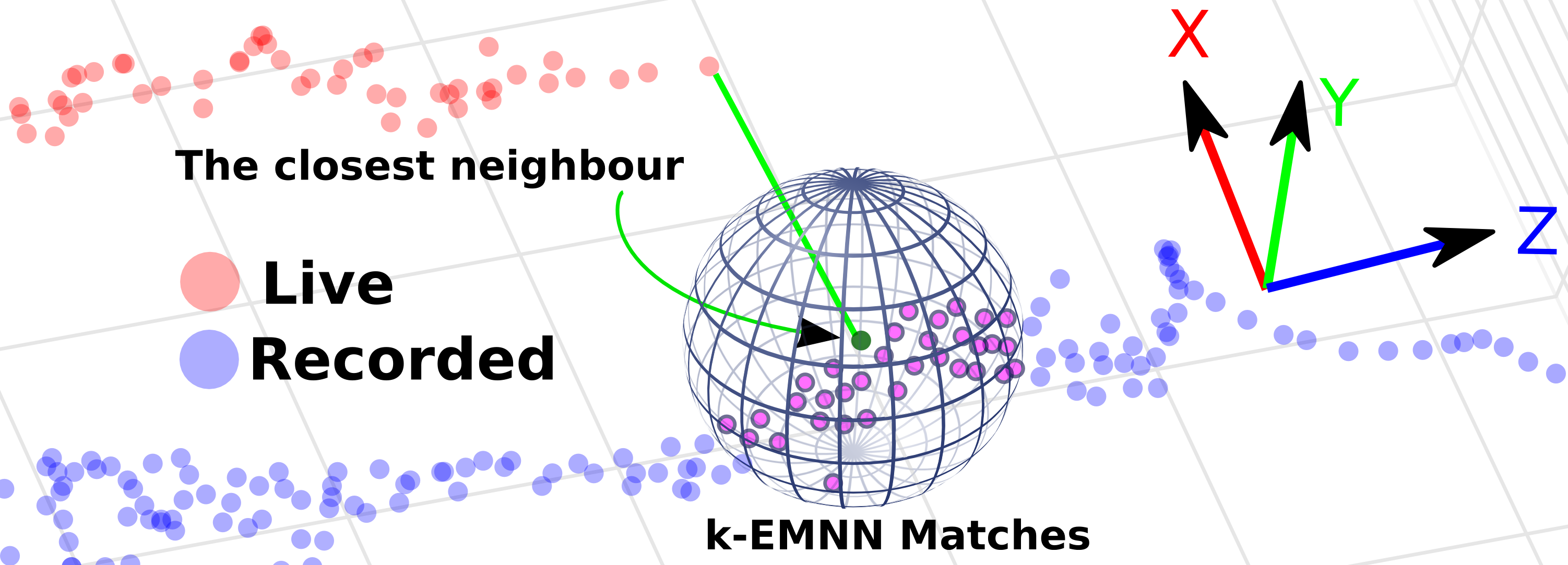
In an earlier paper, we introduced the first approach to inter-operative relocalization using electromagnetic tracking system. In [21] , we propose three incremental experiments to our approach. First, we analyse the error bounds of our system on synthetic data with a realistic noise model. Second, we provide a pseudo ground-truth on in-vivo pig data using an optical tracking system. Accuracy results obtained were consistent with the synthetic experiments despite uncertainty introduced due to breathing motion, and remain inside acceptable error margins according to medical experts. Finally, a third experiment was designed using data from pigs to simulate a real task of biopsy site relocalization, and evaluated by ten experts. It clearly demonstrated the benefit of our system towards assisted guidance by improving the biopsy site retrieval rate from 47.5% to 94%.
This inter-operative relocalization framework was then further extended in [53] to provide a constrained image based search as shown in Fig. 2 to obtain the best view point match to the live view. Within this context, we investigate the effect of (a) the choice of feature descriptors and colour-space, (b) filtering of uninformative frames and (c) endoscopic modality, for view point localization. Our experiments indicate an improvement in the best view-point retrieval rate to [92%, 87%] from [73%, 76%] (in our previous approach) for Narrow band imaging and white-light endoscopic image modalities.
|
Segmentation and anatomic variability of the cochlea and other temporal bone structures from medical images
Participants : Thomas Demarcy [Correspondent] , Hervé Delingette, Clair Vandersteen [IUFC, Nice] , Dan Gnansia [Oticon Medical] , Nicholas Ayache.
This work is supported by the National Association for Research in Technology (ANRT) through the CIFRE Grant 2013-1165 and Oticon Medical (Vallauris). Part of this work is also funded by the European Research Council through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images). This work is done in collaboration with the Department of Ear Nose Throat Surgery (IUFC, Nice) and the Nice University Hospital (CHU).
image segmentation ; surgery planning ; shape modelling ; anatomic variability ; cochlear implant ; temporal bone
-
We designed a parametric shape model of the intracochlear anatomy with anatomical prior learned from temporal bones high-resolution images, see Fig. 3 .
-
We evaluated the cochleostomy location regarding two surgical approaches (endaural compared to conventional posterior tympanotomy) [20] .
|
Structured sparse Bayesian modelling for non-rigid registration and cardiac motion tracking
Participants : Loic Le Folgoc [Correspondent] , Hervé Delingette, Antonio Criminisi, Nicholas Ayache.
This work has been partly supported by the Inria – Microsoft Research Joint Center and by the European Research Council through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Non-rigid Registration - Structured Sparse Bayesian Learning - Automatic Relevance Determination - Reversible-jump Markov Chain Monte Carlo - Cardiac Motion Tracking - Uncertainty Quantification
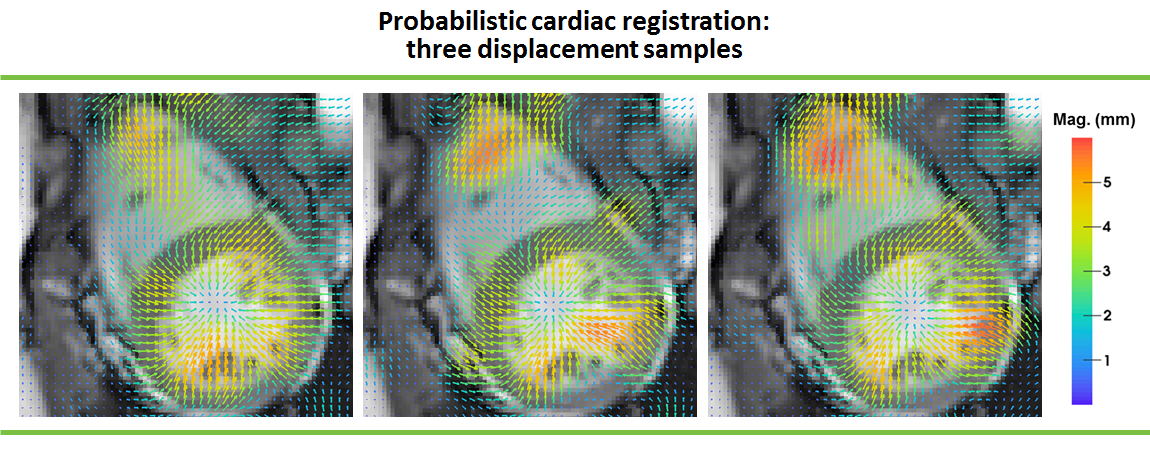
We developed a generic structured sparse Bayesian model of image registration with three main contributions: an extended image similarity term, the automated tuning of registration parameters and uncertainty quantification. We proposed an approximate inference scheme that is tractable on 4D clinical data. We demonstrated the performance of our approach on cine MR, tagged MR and 3D Ultra Sound cardiac images, and showed state-of-the-art results on benchmark datasets evaluating accuracy of motion and strain.
Moreover, we evaluated the quality of uncertainty estimates returned by the approximate inference scheme. We compare the predictions of the approximate scheme with those of an inference scheme developed on the grounds of reversible jump Markov Chain Monte-Carlo [94] (see Fig. 4 ). We provided more insight into the theoretical properties of the sparse structured Bayesian model and into the empirical behaviour of both inference schemes.
This work is described in the PhD manuscript of Loïc Le Folgoc, defended at Université Nice Sophia Antipolis, 2015 [6] .
|
Image Segmentation and Synthesis of brain tumor MR images
Participants : Nicolas Cordier [correspondent] , Hervé Delingette, Nicholas Ayache.
Part of this work was funded by the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Brain, MRI, Glioma, Patch-based Segmentation, Image synthesis
The segmentation of glioblastoma, the most severe case of brain tumors, is a crucial step for diagnostic assessment and therapy planning. In order to perform the manual delineation of the tumor compartments, the clinicians have to concurrently screen multi-channel 3D MRI, which makes the process both time-consuming and subject to inter-expert delineation variability.
We have developed 2 contributions for the analysis of MR brain tumor images:
-
A patch-based multi-atlas automatic glioma segmentation algorithm[13] . Unlike prior work on patch-based multi-atlas segmentation, our approach does not assume any prior knowledge about the location of pathological structures (no local search window).
-
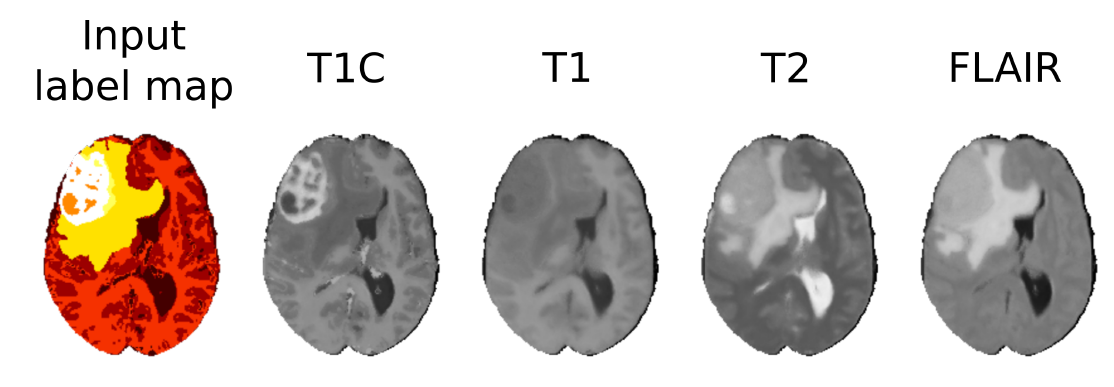
A patch-based image synthesis algorithm (see Fig.5 ) [4] , which generates multi-sequence MR images of the brain with glioma from a single label image. The synthesis of images may be useful to benchmark segmentation algorithms or to increase the size of annotated medical image databases.
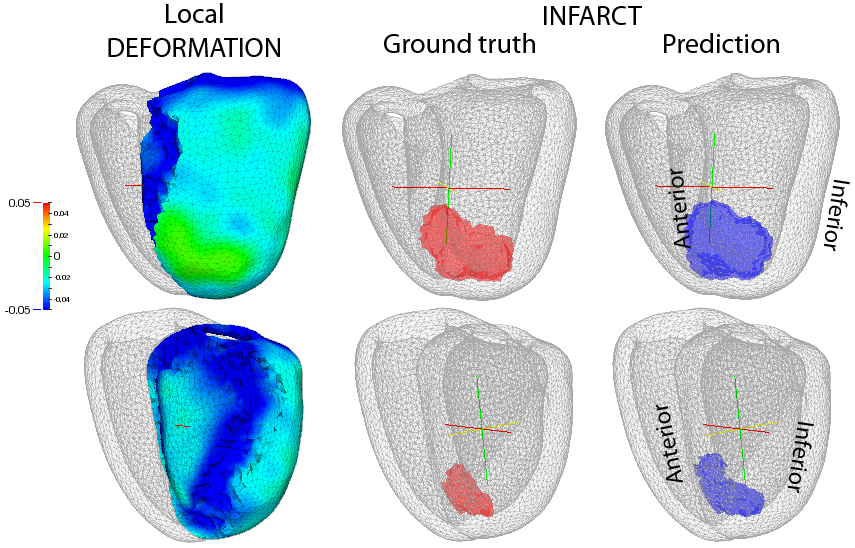
Infarct localization from myocardial deformation
Participants : Nicolas Duchateau [Correspondent] , Maxime Sermesant.
This work received the partial support from the European Union 7th Framework Programme (VP2HF FP7-2013-611823) and the European Research Council (MedYMA ERC-AdG-2011-291080).
Myocardial infarct, Computer-aided diagnosis, Dimensionality reduction, Biomechanical modeling
-
We investigate new methods for predicting the location of myocardial infarcts from local wall deformation [31] , which is useful for risk stratification from routine examinations such as 3D echocardiography. In a broader perspective, this project also aims at determining relevant biomarkers to study cardiac function [54] , and eventually at combining several of those markers in an efficient manner [59] .
-
Non-linear dimensionality reduction aims at estimating the Euclidean space of coordinates encoding deformation patterns, and is combined with multi-scale kernel regressions to infer the low-dimensional coordinates and the infarct location of new cases.
-
These concepts were tested on 500 synthetic cases with infarcts of random extent, shape, and location, generated from a realistic electromechanical model. Our prediction goes beyond the current diagnosis of infarct either achieved at the global or segmental level, and significantly outperforms the clinically-used thresholding of the deformation patterns.